Shen Y, Khanna R, Carle CM, Quail PH. Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Genetic model for light signaling circa 2000. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Over the past few years, a number of X-ray and NMR structures for bacterial phytochromes and LOV domains have been obtained, and have assisted in clarifying how these photoreceptors work (Harper et al., 2004; Rockwell et al., 2006; Chen et al., 2007; Wagner et al., 2007; Cornilescu et al., 2008; Essen et al., 2008; Ogura et al., 2008; Tokutomi et al., 2008; Yang et al., 2008; Hitomi et al., 2009). There is irony in this story. Chen M. Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Dr Siegelman sat me down with his old notebooks and showed me many puzzling results that scientists could not explain from the 1960s observations that could not be explained by the action of a single photoreceptor acting in a linear signaling pathway. Monte E, Al-Sady B, Leivar P, Quail PH. Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. PIF family members control light-regulated gene expression. The extensive set of mutants for Arabidopsis light, disease and hormone responses will be put to good use over the next decade to understand and test hypotheses about fitness trade-offs during evolution of plants growing in specific environments. Alonso JM, Ecker JR. Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Rockwell NC, Su YS, Lagarias JC. Identification of Arabidopsis cryptochrome mutants enabled identification of the many functions for cryptochromes in plants, including its roles in photoperiodism and flowering, seedling emergence, and as an input to the circadian clock. In vitro kinase assays using phytochrome A identified other substrates of PHYA, including CRY1 and CRY2, and some newly discovered signaling proteins (Fankhauser et al., 1999; Fankhauser, 2000). To date, the photochemistry of the cryptochromes has proven difficult to crack, although the prevalent idea is that cryptochromes act through some sort of redoxdriven reaction. Arabidopsis dominated light-signaling research during the 1990s (Figure 1b). Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. identified the first binding partner of CRY2, a nuclear protein called CIB1 that binds to G-boxes in the promoters of light-regulated genes (Liu et al., 2008). This triggers a transcriptional cascade, leading to the regulated expression of thousands of nuclear genes. For me, one of these incidents occurred in 1987, after I had just given my first job seminar. Phytochrome, a red/far-red light photoreceptor, thus became the first plant receptor whose identity was known at the molecular level. Conformational changes in a photosensory LOV domain monitored by time-resolved NMR spectroscopy. Identification of this class of mutants added an important regulatory component to the pathway shown in Figure 2. Barton KA, Binns AN, Matzke AJ, Chilton MD. Lazarova GI, Kerckhoffs LH, Brandstadter J, Matsui M, Kendrick RE, Cordonnier-Pratt MM, Pratt LH. (in press), Wu Y., Hiratsuka K., Neuhaus G. and Chua N-H. Plant J. The first is that phytochromes are light-regulated protein kinases. complex physiological and developmental responses. With the discovery of cyanobacterial phytochromes in the mid-1990s, Yeh et al. Furthermore, each staff speaks at least 3 or 4 languages, including English, Italian and French. Maloof JN, Borevitz JO, Dabi T, et al. official website and that any information you provide is encrypted was routine. Photoreceptors in plant photomorphogenesis to date.
National Library of Medicine Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. I have retold events as I recall them, but bear in mind that my memories may be distorted by more than 20 years of time. The ease and power of Arabidopsis transformation allows over- or under-expression of specific genes in various mutant backgrounds, thereby conferring unique phenotypes. from specific wavelengths of light may be amplified and coordinated, resulting in Elaine Tobins lab weighed in with the significant finding that light acts through phytochrome to regulate gene expression in plants (Silverthorne and Tobin, 1984). The hostel is organized, clean and gives value for money. Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Light-regulated transcriptional networks in higher plants. Kebrom TH, Brutnell TP. were able to show quite clearly that bacterial phytochromes are light-regulated kinases (Yeh et al., 1997). The subcellular localization of phytochrome itself is light-regulated. PHYA, as well as downstream effectors, e.g. The hostel is safe and has friendly staff. Definitely yes. The speaker was Harold William Siegelman, an icon in the phytochrome field, who, in the 1960s, not only gave the intriguing photoperiodic pigment its name, but also partially purified holo-phytochrome and visualized the bluegreen pigment in a cuvette (Butler et al., 1959; Siegelman et al., 1966). Light regulation of plant gene expression by an upstream enhancer-like element. There was great excitement regarding the potential for paradigm-shifting discoveries in plant biology, so much that the US National Science Foundation started a post-doctoral program specifically in the area of plant molecular biology. Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. government site. Copyright 2022 Elsevier Inc. except certain content provided by third parties. Mas P, Devlin PF, Panda S, Kay SA. Mutations in individual phytochromes were characterized, solving some of the dilemmas in Dr Siegelmans notebooks, while at the same time revealing even more complexity in the light-signaling network (reviewed by Neff et al., 2000; Briggs and Olney, 2001). Lam E, Chua N-H. GT-1 binding site confers light responsive expression in transgenic tobacco. The completion of the Arabidopsis genome sequence in 2000 (Arabidopsis Genome Initiative, 2000) has facilitated proteomic studies, design of gene expression and full-genome tiling arrays for global gene expression and epigenetic studies, as well as the positional cloning of genes (Clark et al., 2007; Yazaki et al., 2007; Lister et al., 2009). At the same time, genetic experiments performed in Briggs lab and later in Liscums lab, identified a number of phototropism-defective mutants, nph1, 2, 3 and 4 (Motchoulski and Liscum, 1999; Harper et al., 2000). Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Many of these proteins function in the regulation of protein stability and are members of conserved signaling modules found in both plants and animals. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Dos And Donts Of Packing For A Hotel Stay. Jiao Y, Lau OS, Deng XW. I was with Elliot Meyerowitz, a guest speaker at the first Arabidopsis lab course, and my fellow instructors, Joe Ecker and Sakis Theologis.
Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Dos And Donts Of Packing For A Hotel Stay. Jiao Y, Lau OS, Deng XW. I was with Elliot Meyerowitz, a guest speaker at the first Arabidopsis lab course, and my fellow instructors, Joe Ecker and Sakis Theologis.
Also identified were many positively acting signaling components, including transcription factors, chaperones and scaffolds (reviewed by Neff et al., 2000). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. In addition to controlling phototropism, characterization of phot1 phot2 double mutants implied a role for phototropins in blue light-dependent chloroplast relocation (Kagawa et al., 2001), stomatal opening (Kinoshita et al., 2001) and growth (Folta et al., 2003). Simpson J, Timko MP, Cashmore AR, Schell J, Van Montagu M, Herrera-Estrella L. Light-inducible and tissue-specific expression of a chimaeric gene under control of the 5-flanking sequence of a pea chlorophyll. Were a smart option for all visitors looking for budget accommodation in Lombardy. showed that blue light could activate phosphorylation of a plasma membrane protein from growing regions of etiolated pea seedlings (Gallagher et al., 1988). Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN. Apart from accommodation, we also offer several amenities to make your stay at Hostel Lombardia comfortable and memorable. Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Essen LO, Mailliet J, Hughes J. Strikingly, association testing suggests that the PHYB region is an important determinant of the light response across many A. thaliana accessions (Filiault et al., 2008). Hershey HP, Colbert JT, Lissemore JL, Barker RF, Quail PH. Wertz IE, ORourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. It was still the days of slide projectors, so the lights in the small seminar room were dimmed. These loss-of-function mutations were epistatic to the long-hypocotyl mutants, implying that negative regulators existed downstream from light perception that prevented photomorphogenesis from occurring when no light was available (Chory, 1992). This review is not meant to cover all aspects of light signaling (for instance, I did not even broach the exciting fields of circadian biology, photoperiod and flowering). Arabidopsis DDB1CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. FOIA Ogura Y, Komatsu A, Zikihara K, Nanjo T, Tokutomi S, Wada M, Kiyosue T. Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in. My answer is a resounding no! The role of phytochrome B and simulated shading. Later, Arabidopsis genomic libraries proved useful in identifying a phytochrome gene family (five members in Arabidopsis) (Sharrock and Quail, 1989). The https:// ensures that you are connecting to the We also organize various fun activities for our guests. You will then receive an email that contains a secure link for resetting your password, If the address matches a valid account an email will be sent to __email__ with instructions for resetting your password. Finally, fluorescent phytochromes have allowed the live imaging of internal organs in intact mice (Shu et al., 2009). Geiser M, Weck E, Dorng HP, Werr W, Courage-Tebbe U, Tillmann E, Starlinger P. Genomic clones of a wild-type allele and a transposable element-induced mutant allele of the sucrose synthase gene of. This hotel is situated in Porta Romana with Bocconi University, Fondazione Prada and the University of Milan nearby. Similarly, the human homologs of Arabidopsis COP1 and DET1 proteins were recently shown to be important negative regulators of the human tumor suppressor p53 (Yi and Deng, 2005). Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. The response was rapid (shorter than 15 min) and was universal among the angiosperms. regulate plant development. (in press), 34 Mayer R., Raventos D. and Chua N-H. Plant Cell (in press), DOI: https://doi.org/10.1016/S0962-8924(97)10043-5, If you don't remember your password, you can reset it by entering your email address and clicking the Reset Password button. We also offer discounts and other great promotions from time to time. PHYA is a phosphoprotein in vivo, and at least one serine is phosphorylated in a light-dependent manner. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Murai N, Kemp JD, Sutton DW, et al. I think Bill Siegelman and his mentors would have been proud of where their field has gone. In flies, cryptochromes are both photoreceptors and components of the circadian clock, while, in humans, cryptochromes appear to be solely clock components. Its accessible through the Montenapoleone Fashion District. You know, theres a reason that we all fled from phytochrome research. Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. Weve hosted hundreds of thousands of guests from around the world over the years. Herrera-Estrella L, Block MD, Messens E, Hernalsteens JP, Montagu MV, Schell J. Chimeric genes as dominant selectable markers in plant cells. Genetic and molecular screens, most of which focused on seedling responses, especially those that are part of de-etiolation, identified many dozens of genes acting downstream of photoreceptors. 1983 was an especially good year: the first reports of stable plant transformation were published (Barton et al., 1983; Caplan et al., 1983; Fraley et al., 1983; Herrera-Estrella et al., 1983; Murai et al., 1983), maize transposons were cloned (Burr and Burr, 1982; Geiser et al., 1982; Fedoroff et al., 1983), and an Arabidopsis genetic map with 100 phenotypic markers scattered across the five chromosomes became available (Koornneef et al., 1983). Light signal-transduction pathways are a central component of the mechanisms that COP1 is an E3 ligase that targets both labile photoreceptors, e.g.
Mutation in early signaling intermediates are expected to have a phenotype only under the specific light conditions activating their photoreceptor, and known molecules fitting this requirement include several transcription factors that directly interact with photoreceptors, as well as other molecules that are phosphorylated by phytochromes. and transmitted securely. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. will also be available for a limited time. Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. Dissection of light-signaling networks has had profound implications, not only for all flowering plants, but for human biology as well. Bruce WB, Christensen AH, Klein T, Fromm M, Quail PH. In this review, I provide a brief history and summarize the current state of knowledge of light-signaling research in plants, focusing on how the sequence of Arabidopsis enabled discovery of the principle components and dissection of the molecular mechanisms. Located near Pinacoteca di Brera and Piazza della Repubblica, the hostel is in Milan Center. The content on this site is intended for healthcare professionals. Light: an indicator of time and place. Genetically encoded photoswitching of actin assembly through the Cdc42WASPArp2/3 complex pathway. Light-mediated changes in two proteins found associated with plasma membrane fractions from pea stem sections. Higher-plant phytochromes also have a histidine-kinase related domain, but no histidine kinase activity could be found. The past 30 years has seen a tremendous increase in our understanding of the light-signaling networks of higher plants. Science is truly a collective effort of many individuals who both compete and collaborate with each other in search of the truth. Because all these proteins are regulated by light, oxygen or voltage, the domains were assigned the acronym LOV (Christie et al., 2002; Briggs, 2007). Vierstra RD, Quail PH. Borevitz JO, Maloof JN, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of. Several genes encoding the small subunit of RuBisCO were cloned, as well as genes for the light-harvesting chlorophyll a/b binding proteins. A perfect problem for a geneticist! Yanagawa Y, Sullivan JA, Komatsu S, et al. Expression of bacterial genes in plant cells. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. These domains also show sequence similarity to domains found in a number of signaling proteins in organisms from all kingdoms of life. Maloof JN, Borevitz JO, Weigel D, Chory J. A 30-year timeline of light-signaling research.
Light-mediated changes in two proteins found associated with plasma membrane fractions from pea stem sections. Higher-plant phytochromes also have a histidine-kinase related domain, but no histidine kinase activity could be found. The past 30 years has seen a tremendous increase in our understanding of the light-signaling networks of higher plants. Science is truly a collective effort of many individuals who both compete and collaborate with each other in search of the truth. Because all these proteins are regulated by light, oxygen or voltage, the domains were assigned the acronym LOV (Christie et al., 2002; Briggs, 2007). Vierstra RD, Quail PH. Borevitz JO, Maloof JN, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of. Several genes encoding the small subunit of RuBisCO were cloned, as well as genes for the light-harvesting chlorophyll a/b binding proteins. A perfect problem for a geneticist! Yanagawa Y, Sullivan JA, Komatsu S, et al. Expression of bacterial genes in plant cells. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. These domains also show sequence similarity to domains found in a number of signaling proteins in organisms from all kingdoms of life. Maloof JN, Borevitz JO, Weigel D, Chory J. A 30-year timeline of light-signaling research.
Molecular cloning of cDNA for. Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Mutants clarified the overlapping, and unique, functions of the various phytochromes and led to the discovery of cryptochromes, phototropins and other photoreceptors in plants. El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Yeh KC, Wu SH, Murphy JT, Lagarias JC. Federal government websites often end in .gov or .mil. Combined with the work of Koornneef and colleagues on CRY2 (El-Din El-Assal et al., 2001; El-Assal et al., 2004), and Weinig et al. Model for phytochrome signaling circa 1989. The study of light signaling in plants has not only provided insight into plant growth and development, but has also led to the discovery of conserved proteins that regulate transcription, tumorigenesis and lipid metabolism in metazoans. We target visitors whore looking for short-term or long-term stay at affordable costs. The cloning of NPH1 suggested it to be the photoreceptor for phototropism, as nph1 mutants are impaired in light-activated phosphorylation and phototropism (Christie et al., 1998). Hostel Lombardia offers affordable accommodation to its visitors. LOV domain fusions are also being used to probe conformational changes in proteins (Strickland et al., 2008). A genetically encoded photoactivatable Rac controls the motility of living cells. Ds controlling elements of maize at the shrunken locus are large and dissimilar insertions. In addition, using tissue- and cell type-specific Arabidopsis promoters to express components of light-signaling pathways in a subset of cells is allowing us to understand which portions of a plants response to light are cell autonomous (Tanaka et al., 2002; Endo et al., 2005, 2007; Endo and Nagatani, 2008). Bethesda, MD 20894, Web Policies Because different spectral qualities trigger the same developmental responses using different photoreceptors, it was hypothesized that common late-acting signaling intermediates are used. Lurking in the shadows, a small, insignificant weed was about to burst onto the scene. Linkage map of, Kwok SF, Piekos B, Misra S, Deng X-W. A complement of ten essential and pleiotropic arabidopsis. Weigel D, Mott R. The 1001 genomes project for. However, it lacks kitchen equipment. It was a very exciting time in the light-signaling field as well. The new PMC design is here! This review focuses upon recent However, prices usually go slightly higher during the holiday season such as Christmas and the New Years Eve. Eight of the proteins are members of an evolutionarily conserved complex called the CSN (COP9 signalosome) (Wei et al., 1998). Studying the mechanisms by which plants respond to light has had an impact beyond plants. Its a question, How to choose where to go on a holiday Choosing where to go on a holiday is one of the most challenging decisions. The availability of the fully sequenced and annotated genome of Arabidopsis gave us a snapshot of what makes a plant a plant. Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng X-W. 8600 Rockville Pike Importantly, it outlines how genetic studies led to the identification of photoreceptors and signaling components that are not only relevant in plants, but play key roles in mammals. A few papers, such as Koornneefs paper on long-hypocotyl mutants of Arabidopsis, are notable (Koornneef et al., 1980). My job talk was over and it was time for probing questions, but the first speaker did not ask me for clarification; rather, out of the darkness in the back of the room came the proclamation Why, you are a very brave and naive girl! Coupled with natural variation studies (Weigel and Mott, 2009), and the Arabidopsis molecular genetics toolkit (Lister et al., 2009), the possibilities are limitless. We all love our iPads, but are they bad for the environment?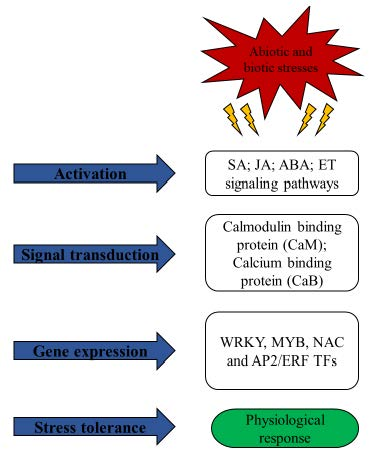 Functional interaction of phytochrome B and cryptochrome 2. Click here to explore this opportunity. Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome BGFP fusion protein to the nucleus in transgenic Arabidopsis. Structurefunction studies on phytochrome. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. The structure of a complete phytochrome sensory module in the Pr ground state. Work in multiple laboratories led to the exciting discovery that light positively regulates transcription of these genes (Simpson et al., 1985; Timko et al., 1985; Gilmartin et al., 1990; Lam and Chua, 1990). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. The LOV domain: a chromophore module servicing multiple photoreceptors. Strickland D, Moffat K, Sosnick TR. Please enter a term before submitting your search. Cloning the Arabidopsis gene allowed predictions of the protein sequence, and enabled the discovery of cryptochromes from both flies and humans (Ceriani et al., 1999). Plant Mol. Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Arabidopsis COP10 forms a complex with DDB1 and DET1, Yang X, Kuk J, Moffat K. Crystal structure of. Genetic control of light-inhibited hypocotyl elongation in, Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. They keep on coming back to us each time they visit Lombardy. Other hostels in Lombardy include Combo Milano, Milano Ostello, Hostel Colours, Central Hostel BG, Ostello del Castello Tirano, Milan Hotel, and Ostello La Goliarda. Levskaya A, Weiner OD, Lim WA, Voigt CA. However, CRY1 has no photolyase activity. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex.
Functional interaction of phytochrome B and cryptochrome 2. Click here to explore this opportunity. Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome BGFP fusion protein to the nucleus in transgenic Arabidopsis. Structurefunction studies on phytochrome. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. The structure of a complete phytochrome sensory module in the Pr ground state. Work in multiple laboratories led to the exciting discovery that light positively regulates transcription of these genes (Simpson et al., 1985; Timko et al., 1985; Gilmartin et al., 1990; Lam and Chua, 1990). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. The LOV domain: a chromophore module servicing multiple photoreceptors. Strickland D, Moffat K, Sosnick TR. Please enter a term before submitting your search. Cloning the Arabidopsis gene allowed predictions of the protein sequence, and enabled the discovery of cryptochromes from both flies and humans (Ceriani et al., 1999). Plant Mol. Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Arabidopsis COP10 forms a complex with DDB1 and DET1, Yang X, Kuk J, Moffat K. Crystal structure of. Genetic control of light-inhibited hypocotyl elongation in, Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. They keep on coming back to us each time they visit Lombardy. Other hostels in Lombardy include Combo Milano, Milano Ostello, Hostel Colours, Central Hostel BG, Ostello del Castello Tirano, Milan Hotel, and Ostello La Goliarda. Levskaya A, Weiner OD, Lim WA, Voigt CA. However, CRY1 has no photolyase activity. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. 
National Library of Medicine Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. I have retold events as I recall them, but bear in mind that my memories may be distorted by more than 20 years of time. The ease and power of Arabidopsis transformation allows over- or under-expression of specific genes in various mutant backgrounds, thereby conferring unique phenotypes. from specific wavelengths of light may be amplified and coordinated, resulting in Elaine Tobins lab weighed in with the significant finding that light acts through phytochrome to regulate gene expression in plants (Silverthorne and Tobin, 1984). The hostel is organized, clean and gives value for money. Hitomi K, DiTacchio L, Arvai AS, Yamamoto J, Kim ST, Todo T, Tainer JA, Iwai S, Panda S, Getzoff ED. Light-regulated transcriptional networks in higher plants. Kebrom TH, Brutnell TP. were able to show quite clearly that bacterial phytochromes are light-regulated kinases (Yeh et al., 1997). The subcellular localization of phytochrome itself is light-regulated. PHYA, as well as downstream effectors, e.g. The hostel is safe and has friendly staff. Definitely yes. The speaker was Harold William Siegelman, an icon in the phytochrome field, who, in the 1960s, not only gave the intriguing photoperiodic pigment its name, but also partially purified holo-phytochrome and visualized the bluegreen pigment in a cuvette (Butler et al., 1959; Siegelman et al., 1966). Light regulation of plant gene expression by an upstream enhancer-like element. There was great excitement regarding the potential for paradigm-shifting discoveries in plant biology, so much that the US National Science Foundation started a post-doctoral program specifically in the area of plant molecular biology. Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. government site. Copyright 2022 Elsevier Inc. except certain content provided by third parties. Mas P, Devlin PF, Panda S, Kay SA. Mutations in individual phytochromes were characterized, solving some of the dilemmas in Dr Siegelmans notebooks, while at the same time revealing even more complexity in the light-signaling network (reviewed by Neff et al., 2000; Briggs and Olney, 2001). Lam E, Chua N-H. GT-1 binding site confers light responsive expression in transgenic tobacco. The completion of the Arabidopsis genome sequence in 2000 (Arabidopsis Genome Initiative, 2000) has facilitated proteomic studies, design of gene expression and full-genome tiling arrays for global gene expression and epigenetic studies, as well as the positional cloning of genes (Clark et al., 2007; Yazaki et al., 2007; Lister et al., 2009). At the same time, genetic experiments performed in Briggs lab and later in Liscums lab, identified a number of phototropism-defective mutants, nph1, 2, 3 and 4 (Motchoulski and Liscum, 1999; Harper et al., 2000). Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Many of these proteins function in the regulation of protein stability and are members of conserved signaling modules found in both plants and animals.
 Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Dos And Donts Of Packing For A Hotel Stay. Jiao Y, Lau OS, Deng XW. I was with Elliot Meyerowitz, a guest speaker at the first Arabidopsis lab course, and my fellow instructors, Joe Ecker and Sakis Theologis.
Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Dos And Donts Of Packing For A Hotel Stay. Jiao Y, Lau OS, Deng XW. I was with Elliot Meyerowitz, a guest speaker at the first Arabidopsis lab course, and my fellow instructors, Joe Ecker and Sakis Theologis. Also identified were many positively acting signaling components, including transcription factors, chaperones and scaffolds (reviewed by Neff et al., 2000). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. In addition to controlling phototropism, characterization of phot1 phot2 double mutants implied a role for phototropins in blue light-dependent chloroplast relocation (Kagawa et al., 2001), stomatal opening (Kinoshita et al., 2001) and growth (Folta et al., 2003). Simpson J, Timko MP, Cashmore AR, Schell J, Van Montagu M, Herrera-Estrella L. Light-inducible and tissue-specific expression of a chimaeric gene under control of the 5-flanking sequence of a pea chlorophyll. Were a smart option for all visitors looking for budget accommodation in Lombardy. showed that blue light could activate phosphorylation of a plasma membrane protein from growing regions of etiolated pea seedlings (Gallagher et al., 1988). Filiault DL, Wessinger CA, Dinneny JR, Lutes J, Borevitz JO, Weigel D, Chory J, Maloof JN. Apart from accommodation, we also offer several amenities to make your stay at Hostel Lombardia comfortable and memorable. Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Essen LO, Mailliet J, Hughes J. Strikingly, association testing suggests that the PHYB region is an important determinant of the light response across many A. thaliana accessions (Filiault et al., 2008). Hershey HP, Colbert JT, Lissemore JL, Barker RF, Quail PH. Wertz IE, ORourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. It was still the days of slide projectors, so the lights in the small seminar room were dimmed. These loss-of-function mutations were epistatic to the long-hypocotyl mutants, implying that negative regulators existed downstream from light perception that prevented photomorphogenesis from occurring when no light was available (Chory, 1992). This review is not meant to cover all aspects of light signaling (for instance, I did not even broach the exciting fields of circadian biology, photoperiod and flowering). Arabidopsis DDB1CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. FOIA Ogura Y, Komatsu A, Zikihara K, Nanjo T, Tokutomi S, Wada M, Kiyosue T. Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in. My answer is a resounding no! The role of phytochrome B and simulated shading. Later, Arabidopsis genomic libraries proved useful in identifying a phytochrome gene family (five members in Arabidopsis) (Sharrock and Quail, 1989). The https:// ensures that you are connecting to the We also organize various fun activities for our guests. You will then receive an email that contains a secure link for resetting your password, If the address matches a valid account an email will be sent to __email__ with instructions for resetting your password. Finally, fluorescent phytochromes have allowed the live imaging of internal organs in intact mice (Shu et al., 2009). Geiser M, Weck E, Dorng HP, Werr W, Courage-Tebbe U, Tillmann E, Starlinger P. Genomic clones of a wild-type allele and a transposable element-induced mutant allele of the sucrose synthase gene of. This hotel is situated in Porta Romana with Bocconi University, Fondazione Prada and the University of Milan nearby. Similarly, the human homologs of Arabidopsis COP1 and DET1 proteins were recently shown to be important negative regulators of the human tumor suppressor p53 (Yi and Deng, 2005). Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. The response was rapid (shorter than 15 min) and was universal among the angiosperms. regulate plant development. (in press), 34 Mayer R., Raventos D. and Chua N-H. Plant Cell (in press), DOI: https://doi.org/10.1016/S0962-8924(97)10043-5, If you don't remember your password, you can reset it by entering your email address and clicking the Reset Password button. We also offer discounts and other great promotions from time to time. PHYA is a phosphoprotein in vivo, and at least one serine is phosphorylated in a light-dependent manner. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Murai N, Kemp JD, Sutton DW, et al. I think Bill Siegelman and his mentors would have been proud of where their field has gone. In flies, cryptochromes are both photoreceptors and components of the circadian clock, while, in humans, cryptochromes appear to be solely clock components. Its accessible through the Montenapoleone Fashion District. You know, theres a reason that we all fled from phytochrome research. Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. Weve hosted hundreds of thousands of guests from around the world over the years. Herrera-Estrella L, Block MD, Messens E, Hernalsteens JP, Montagu MV, Schell J. Chimeric genes as dominant selectable markers in plant cells. Genetic and molecular screens, most of which focused on seedling responses, especially those that are part of de-etiolation, identified many dozens of genes acting downstream of photoreceptors. 1983 was an especially good year: the first reports of stable plant transformation were published (Barton et al., 1983; Caplan et al., 1983; Fraley et al., 1983; Herrera-Estrella et al., 1983; Murai et al., 1983), maize transposons were cloned (Burr and Burr, 1982; Geiser et al., 1982; Fedoroff et al., 1983), and an Arabidopsis genetic map with 100 phenotypic markers scattered across the five chromosomes became available (Koornneef et al., 1983). Light signal-transduction pathways are a central component of the mechanisms that COP1 is an E3 ligase that targets both labile photoreceptors, e.g.
Mutation in early signaling intermediates are expected to have a phenotype only under the specific light conditions activating their photoreceptor, and known molecules fitting this requirement include several transcription factors that directly interact with photoreceptors, as well as other molecules that are phosphorylated by phytochromes. and transmitted securely. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. will also be available for a limited time. Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. Dissection of light-signaling networks has had profound implications, not only for all flowering plants, but for human biology as well. Bruce WB, Christensen AH, Klein T, Fromm M, Quail PH. In this review, I provide a brief history and summarize the current state of knowledge of light-signaling research in plants, focusing on how the sequence of Arabidopsis enabled discovery of the principle components and dissection of the molecular mechanisms. Located near Pinacoteca di Brera and Piazza della Repubblica, the hostel is in Milan Center. The content on this site is intended for healthcare professionals. Light: an indicator of time and place. Genetically encoded photoswitching of actin assembly through the Cdc42WASPArp2/3 complex pathway.
 Light-mediated changes in two proteins found associated with plasma membrane fractions from pea stem sections. Higher-plant phytochromes also have a histidine-kinase related domain, but no histidine kinase activity could be found. The past 30 years has seen a tremendous increase in our understanding of the light-signaling networks of higher plants. Science is truly a collective effort of many individuals who both compete and collaborate with each other in search of the truth. Because all these proteins are regulated by light, oxygen or voltage, the domains were assigned the acronym LOV (Christie et al., 2002; Briggs, 2007). Vierstra RD, Quail PH. Borevitz JO, Maloof JN, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of. Several genes encoding the small subunit of RuBisCO were cloned, as well as genes for the light-harvesting chlorophyll a/b binding proteins. A perfect problem for a geneticist! Yanagawa Y, Sullivan JA, Komatsu S, et al. Expression of bacterial genes in plant cells. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. These domains also show sequence similarity to domains found in a number of signaling proteins in organisms from all kingdoms of life. Maloof JN, Borevitz JO, Weigel D, Chory J. A 30-year timeline of light-signaling research.
Light-mediated changes in two proteins found associated with plasma membrane fractions from pea stem sections. Higher-plant phytochromes also have a histidine-kinase related domain, but no histidine kinase activity could be found. The past 30 years has seen a tremendous increase in our understanding of the light-signaling networks of higher plants. Science is truly a collective effort of many individuals who both compete and collaborate with each other in search of the truth. Because all these proteins are regulated by light, oxygen or voltage, the domains were assigned the acronym LOV (Christie et al., 2002; Briggs, 2007). Vierstra RD, Quail PH. Borevitz JO, Maloof JN, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of. Several genes encoding the small subunit of RuBisCO were cloned, as well as genes for the light-harvesting chlorophyll a/b binding proteins. A perfect problem for a geneticist! Yanagawa Y, Sullivan JA, Komatsu S, et al. Expression of bacterial genes in plant cells. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. These domains also show sequence similarity to domains found in a number of signaling proteins in organisms from all kingdoms of life. Maloof JN, Borevitz JO, Weigel D, Chory J. A 30-year timeline of light-signaling research. Molecular cloning of cDNA for. Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Mutants clarified the overlapping, and unique, functions of the various phytochromes and led to the discovery of cryptochromes, phototropins and other photoreceptors in plants. El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Yeh KC, Wu SH, Murphy JT, Lagarias JC. Federal government websites often end in .gov or .mil. Combined with the work of Koornneef and colleagues on CRY2 (El-Din El-Assal et al., 2001; El-Assal et al., 2004), and Weinig et al. Model for phytochrome signaling circa 1989. The study of light signaling in plants has not only provided insight into plant growth and development, but has also led to the discovery of conserved proteins that regulate transcription, tumorigenesis and lipid metabolism in metazoans. We target visitors whore looking for short-term or long-term stay at affordable costs. The cloning of NPH1 suggested it to be the photoreceptor for phototropism, as nph1 mutants are impaired in light-activated phosphorylation and phototropism (Christie et al., 1998). Hostel Lombardia offers affordable accommodation to its visitors. LOV domain fusions are also being used to probe conformational changes in proteins (Strickland et al., 2008). A genetically encoded photoactivatable Rac controls the motility of living cells. Ds controlling elements of maize at the shrunken locus are large and dissimilar insertions. In addition, using tissue- and cell type-specific Arabidopsis promoters to express components of light-signaling pathways in a subset of cells is allowing us to understand which portions of a plants response to light are cell autonomous (Tanaka et al., 2002; Endo et al., 2005, 2007; Endo and Nagatani, 2008). Bethesda, MD 20894, Web Policies Because different spectral qualities trigger the same developmental responses using different photoreceptors, it was hypothesized that common late-acting signaling intermediates are used. Lurking in the shadows, a small, insignificant weed was about to burst onto the scene. Linkage map of, Kwok SF, Piekos B, Misra S, Deng X-W. A complement of ten essential and pleiotropic arabidopsis. Weigel D, Mott R. The 1001 genomes project for. However, it lacks kitchen equipment. It was a very exciting time in the light-signaling field as well. The new PMC design is here! This review focuses upon recent However, prices usually go slightly higher during the holiday season such as Christmas and the New Years Eve. Eight of the proteins are members of an evolutionarily conserved complex called the CSN (COP9 signalosome) (Wei et al., 1998). Studying the mechanisms by which plants respond to light has had an impact beyond plants. Its a question, How to choose where to go on a holiday Choosing where to go on a holiday is one of the most challenging decisions. The availability of the fully sequenced and annotated genome of Arabidopsis gave us a snapshot of what makes a plant a plant. Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng X-W. 8600 Rockville Pike Importantly, it outlines how genetic studies led to the identification of photoreceptors and signaling components that are not only relevant in plants, but play key roles in mammals. A few papers, such as Koornneefs paper on long-hypocotyl mutants of Arabidopsis, are notable (Koornneef et al., 1980). My job talk was over and it was time for probing questions, but the first speaker did not ask me for clarification; rather, out of the darkness in the back of the room came the proclamation Why, you are a very brave and naive girl! Coupled with natural variation studies (Weigel and Mott, 2009), and the Arabidopsis molecular genetics toolkit (Lister et al., 2009), the possibilities are limitless. We all love our iPads, but are they bad for the environment?
 Functional interaction of phytochrome B and cryptochrome 2. Click here to explore this opportunity. Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome BGFP fusion protein to the nucleus in transgenic Arabidopsis. Structurefunction studies on phytochrome. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. The structure of a complete phytochrome sensory module in the Pr ground state. Work in multiple laboratories led to the exciting discovery that light positively regulates transcription of these genes (Simpson et al., 1985; Timko et al., 1985; Gilmartin et al., 1990; Lam and Chua, 1990). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. The LOV domain: a chromophore module servicing multiple photoreceptors. Strickland D, Moffat K, Sosnick TR. Please enter a term before submitting your search. Cloning the Arabidopsis gene allowed predictions of the protein sequence, and enabled the discovery of cryptochromes from both flies and humans (Ceriani et al., 1999). Plant Mol. Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Arabidopsis COP10 forms a complex with DDB1 and DET1, Yang X, Kuk J, Moffat K. Crystal structure of. Genetic control of light-inhibited hypocotyl elongation in, Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. They keep on coming back to us each time they visit Lombardy. Other hostels in Lombardy include Combo Milano, Milano Ostello, Hostel Colours, Central Hostel BG, Ostello del Castello Tirano, Milan Hotel, and Ostello La Goliarda. Levskaya A, Weiner OD, Lim WA, Voigt CA. However, CRY1 has no photolyase activity. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex.
Functional interaction of phytochrome B and cryptochrome 2. Click here to explore this opportunity. Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome BGFP fusion protein to the nucleus in transgenic Arabidopsis. Structurefunction studies on phytochrome. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. The structure of a complete phytochrome sensory module in the Pr ground state. Work in multiple laboratories led to the exciting discovery that light positively regulates transcription of these genes (Simpson et al., 1985; Timko et al., 1985; Gilmartin et al., 1990; Lam and Chua, 1990). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Primary inhibition of hypocotyl growth and phototropism depend differently on phototropin-mediated increases in cytoplasmic calcium induced by blue light. The LOV domain: a chromophore module servicing multiple photoreceptors. Strickland D, Moffat K, Sosnick TR. Please enter a term before submitting your search. Cloning the Arabidopsis gene allowed predictions of the protein sequence, and enabled the discovery of cryptochromes from both flies and humans (Ceriani et al., 1999). Plant Mol. Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. Arabidopsis COP10 forms a complex with DDB1 and DET1, Yang X, Kuk J, Moffat K. Crystal structure of. Genetic control of light-inhibited hypocotyl elongation in, Koornneef M, van Eden J, Hanhart CJ, Stam P, Braaksma FJ, Feenstra WJ. They keep on coming back to us each time they visit Lombardy. Other hostels in Lombardy include Combo Milano, Milano Ostello, Hostel Colours, Central Hostel BG, Ostello del Castello Tirano, Milan Hotel, and Ostello La Goliarda. Levskaya A, Weiner OD, Lim WA, Voigt CA. However, CRY1 has no photolyase activity. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. 