The user selects databases for the file preparation and probe specificity checks.
Thank you for submitting a comment on this article. Us, 2015 - Copyright Eurofins MWG Operon LLC A Eurofins Genomics Company, - TRUEmer The number of sequences and their share of the entire data pool are listed.
In summary, the final set of primers or probes strongly depends on the selected models and parameters. the % GC Content, melting temp (TM), and the reverse complement of the oligo sequence Areas of 4 or more contiguous This software is distributed in the hope that it will be useful to the field of epigenetic bioinformatics, you enter. Oli2go is freely accessible to all users at http://oli2go.ait.ac.at/. but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. Long-Read Sequencing Identifies the First Retrotransposon Insertion and Resolves Structural Variants Causing Antithrombin Deficiency. Our handy Oligo Analysis Tool calculates molecular weight (MW), extinction coefficient 6. In order to maximize the utilization of the server resources, most of the workflow steps are running in parallel using multithreading. OWNERS OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY Oli2go has been successfully tested for the design of an 45-plex assay targeting antibiotic resistance genes. PrimerROC Analysis for Dimer Predictions, "Multiplex bisulfite PCR resequencing of clinical FFPE DNA", Enhancements and modifications of primer design program Primer3, "Nucleic acid duplex stability: influence of base composition on cation effects", "Multiplex polymerase chain reaction: a practical approach", http://www.girinst.org/repbase/index.html. Clozapine's multiple cellular mechanisms: What do we know after more than fifty years? Please note, in either input options, input primer pairs must be listed in the order below: Primer Dimer may output the results in two different formats: The PrimerPlex module is available at www.primer-plex.com. Syntheses and spectral properties of some deazaadenylyl-deazaadenosines (dinucleoside monophosphates with unusual CD spectrum) and closely related dinucleoside monophosphates. Funding for open access charge:H2020 [634137]. - About By default, Primer Suite has the bisulfite primer design function selected on start up. Primer Suite is a web-based primer-design application which addresses these limitations and much more.
The selection of primers and probes starts with the creation of k-mers, ranging from the minimum user-defined primer and probe size to the maximum one, using a step size of 1. The software workflow runs on a Linux server (64 CPUs, 256GB RAM). Sequencing, Gene As Primer3 can only handle one sequence, this step has to be done for each target at least once. Due to the highly positive outcome of this test and the increasing demand for automated multiplex oligonucleotide design solutions, we are consistently working on further improvements of oli2go. This is composed of three different modules: We request that the use of any primers generated through PrimerSuite be cited as following: However primer design for bisulfite The developers of Primer Suite can be contacted via email at jennifer.lu@uqconnect.edu.au. Oligonucleotides are accepted if their secondary structure Tm and G value are below the user-defined thresholds. The results of each probe need to be checked individually by the user. Please check for further notifications by email. Rep.7, 41328; doi: 10.1038/srep41328(2017). 5. Combining the recommendations from different publications, the following factors need to be addressed during the design process: oligonucleotide length and melting temperature (Tm), primer pairing, amplification product size, secondary structure formation, sensitivity and specificity of subsequent reactions (14). PrimerNucleosome PrimerDimer can be accessed via www.primer-dimer.com. Us, - Contact The main software packages used for the implementation are BLAST 2.7.0+, ntthal (which is part of Primer3 2.3.7), BWA, and Python 2.7 together with the Biopython library (21). Subsequently, a primer dimer check is performed using all primers produced in the multiplex design. De Novo design of potential inhibitors against SARS-CoV-2 Mpro. Specific forward and reverse primer pairs resulting from the preceding design task form the input for this last workflow step. button to order directly from this window. 17. The design process involves a number of steps, which use specific parameters to produce high quality oligonucleotides. All rights reserved. For full access to this pdf, sign in to an existing account, or purchase an annual subscription. Oli2go has high potential in improving assay design of oligonucleotide-based experiments. Citing PrimerSuite These primers are checked against all other possible primers of the other input sequences. (A) Illustrates the workflow starting with the input of n DNA sequences, followed by the multiplex design, which is performed independently for each input sequence. Sequences from the Comprehensive Antibiotic Resistance Database (CARD) were used as input for the probe and primer design (24). Version History Furthermore, oli2go significantly reduces the number of manual, error prone steps. Oligos, DNA The design process involves several steps, which use specific parameters to produce oligonucleotides for conducting high quality experiments. It starts with the manual input of target sequences to Primer3. regions simultaneously using minimal amounts of template DNA[1]. Further information can be obtained by emailing us at jennifer.lu@uqconnect.edu.au or via the FAQs section in PrimerSuite resources. PrimerROC Analysis for Dimer predictions, 7. o The FragCalc module is freely available at www.primer-suite.com/fragcalc. enter a second sequence into the Plot Sequence box and click the CALCULATE button However, it should be noted that the enthalpy change upon primer binding is sequence dependent. Oligonucleotides are short, single-stranded nucleic acids used for an increasing variety of biological experiments. LncRNA CTBP1-DT-encoded microprotein DDUP sustains DNA damage response signalling to trigger dual DNA repair mechanisms, Exploration of DNA processing features unravels novel properties of ICE conjugation in Gram-positive bacteria, Role of EXO1 nuclease activity in genome maintenance, the immune response and tumor suppression in, Cap-independent translation and a precisely located RNA sequence enable SARS-CoV-2 to control host translation and escape anti-viral response, Whole-genome long-read TAPS deciphers DNA methylation patterns at base resolution using PacBio SMRT sequencing technology, Chemical Biology and Nucleic Acid Chemistry, Gene Regulation, Chromatin and Epigenetics, http://creativecommons.org/licenses/by-nc/4.0/, Receive exclusive offers and updates from Oxford Academic, MAGMA: analysis of two-channel microarrays made easy, antiSMASH 6.0: improving cluster detection and comparison capabilities, DNA-like duplexes with repetitions. Finally, for each input sequence one primer pair forming no cross dimerization with all other sequences is returned. Furthermore, the specificity can only be checked using one background database derived from a relatively small pool of species. o The PrimerPlex interface is freely available at www.primer-plex.com. mouse), but against bacteria, viruses, fungi, invertebrates, plants, protozoa, archaea and sequences from whole genome shotgun sequence projects and environmental samples, at once. Introduction Afterward, the Tm is calculated for each k-mer (16,19). (B) illustrates the manual design workflow. It is known, that computational automation and big data handling, as performed by oli2go, become more and more important for biologists (25). This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (. Also the manual examination of all specificity check results using online BLAST is impossible to do within in a reasonable time. The [long]/[short] ratios of any two region lengths is determined by applying the following equation to fragment size distribution data: (B) Provides more details on the multiplex probe and primer design steps, which involve k-mer selections, Tm calculations, hairpin checks, probe and primer specificity checks as well as probe and primer pairing for each input sequence independently. thermodynamic models in the algorithm, it showed the best performance considering run time, memory usage, and result quality. Several papers describe in detail the selection of optimized parameters for primer and probe design (3,4,17,18). VI. IN NO EVENT SHALL THE COPYRIGHT o The PrimerDimer interface is freely available at www.primer-dimer.com. The table also contains web links to NCBIs online BLAST and Primer-BLAST to perform additional analysis. If the "Genomic" option is selected, Primer Suite will return an excel workbook with two spreadsheets with the result primers. The length of oligonucleotides is a critical factor for biological experiments because specificity and Tm depend on this physical parameter (2). I. Also displayed for your convenience are The arrows in the figure highlight the iterative nature of the oligonucleotide design process which is responsible for the high work load in comparison to the fully automated oli2go workflow. Dependent on the use case, the default parameters should be tuned meaningfully. The workflow of oli2go is illustrated in Figure1. Independent of the use case, the success of oligonucleotide-based experiments strongly depends on an accurate design and appropriate parameter selection. The purpose of this program is to organise primer pairs into different pools for multiplex PCR [5]. Oli2go has already proven its usability and service to the scientific community during a successful testing period of 6 months performed by an external project team (>10 people, >100 input sequences). This allows the design of highly specific oligonucleotides in multiplex applications, which is further assured by performing dimer checks not only on the primers themselves, but in an all-against-all fashion. 4. The Primer Suite software is a free software for researchers. However, none of them covers the whole workflow (Supplementary Table S1). The following subsections describe the main features of each step in detail.
The resulting probes are merged and submitted to online BLAST where the probe specificity check is performed. Furthermore, primer and probe sequences as well as the initial input sequences are available in FASTA format. Download citation | Download article Published by Oxford University Press on behalf of Nucleic Acids Research. compared the most common Tm calculator tools for PCR oligonucleotide design showing that Primer3 Plus and Primer-BLAST offer the most precise algorithms for an accurate Tm prediction (68). For primer design for genomic DNA, it is recommended you adjust the following parameters for optimal results, or alternatively the default values may be used. The probes were tested on a microarray platform resulting in highly specific signals (Supplementary Figure S2). For bisulfite primer design, it is recommended that users adjust the following parameters for optimal results, else the default values may be used (optimal conditions confirmed in the wet lab to produce quality primers). 9. Supplementary Data are available at NAR Online. Australian Institute for BioEngineering and Nanotechnology (AIBN), Centre for Personalised Nanomedicine, Trau Lab, Jennifer Lu and Darren Korbie. Areas of hybridization are marked with an X. Therefore, it is better to match G values instead of Tm (9). Users may choose to either paste their primer sequences in the textarea, or upload a file in FASTA format. Oli2go is an efficient, user friendly, fully automated multiplex oligonucleotide design tool, which performs primer and different hybridization probe designs as well as specificity and cross dimer checks in a single run. Synthesis. The highly responsive user interface is implemented using Bootstrap 3.3.7 and enables the user to use oli2go on almost any device capable of entering the internet via browser ranging from Laptops, Tablets to Smartphones. AIT Austrian Institute of Technology, Center for Health, and Bioresources, Molecular Diagnostics, Muthgasse 11, 1190 Vienna, Austria. 15. Bakhtiarizadeh M.R., Najaf-Panah M.J., Mousapour H., Salami S.A. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A.M. A systematic review and critical assessment of translational mechanisms relevant for innovative strategies in treatment-resistant schizophrenia. Ilie L., Mohamadi H., Golding Geoffrey B., Smyth W.F. Consequently, researchers do no longer have to use multiple tools, each having their own requirements, input formats, and parameters, to find suitable primers and probes for their experiments. Each primer pool is reported in a seperate spreadsheet and is named pool n (primer pool number). To whom correspondence should be addressed. 14. After the visual evaluation of the results, specific primer candidates are merged in a single file and checked for cross dimerization using MFEprimer. Oxford University Press is a department of the University of Oxford. It starts with the input sequence that has fewest specific primers. Comparing the run time of oli2go with an example of a conventional oligonucleotide design workflow, we can prove that oli2go is significantly faster (Figure2). The home page of the web-based tool oli2go is used to upload the input sequences and to specify the design parameters. where: o The PrimerSuite interface is freely available at www.primer-suite.com. (A) shows the run times of oli2go compared to the conventional manual design workflow shown in B). Primer Suite is a suite of freely available online software for generating primers for multiplex bisulfite PCR. Improvement of chemo- and stereoselectivity for phosphorothioate oligonucleotides in capillary electrophoresis by addition of cyclodextrins. MFEprimer performs a primer dimer and specificity check, which implements the nearest-neighbor model to evaluate the binding stability for multiplex reactions, but misses an oligonucleotide design step (16). The sequences have to be provided in FASTA format, either by upload or by using a designated input box. In addition, the primer dimer formation calculation is limited to maximal 50 primers per run. The Author(s) 2018. The success of widely used oligonucleotide-based experiments, ranging from PCR to microarray, strongly depends on an accurate design. This software uses the tables of thermodynamic parameters suggested by SantaLucia to calculate the secondary structure Tm and G value of the most stable duplex (16). Copyright and Licence Each tool differs in the used design pipeline and input parameters. The primer pairing parameter needs to consider the size of the PCR product, a minimal Tm difference between the primers, or matched G values (1,4,9). (C) Visualizes the primer dimer check, where all primers targeting all input sequences, resulting from the preceding multiplex design, are checked for primer dimer formation. 11. The computationally most expensive steps include BLAST analysis for a high number of specificity checks that have to be computed per target sequence (>100). The hairpin check is implemented using Primer3s nucleotide thermodynamic alignment tool ntthal (12). Moreover, hybridization probes must bind exclusively to the target in order to prevent cross-hybridization to non-targets (11). PrecisePrimer performs primer design for PCR primers involving useful pre-set options for different polymerase buffers and batch design, but lack options for probe design, cross dimerization and specificity checks. The output is presented on a separate web-page and includes a table showing the resulting primers and probes, their Tms, product sizes, hairpin Tms, and G values. PrimerSuite: A High-Throughput Web-Based Primer Design Program for Multiplex Bisulfite PCR. Also applications for sequence assembly, such as Kraken, could not be used, as the oligonucleotide sizes are usually too small for the implemented algorithms (22). The primer specificity check is performed running Primer-BLAST on each primer pair separately. Search for other works by this author on: University of Vienna, Faculty of Informatics, Whringer Strae 29, 1090 Vienna, Austria. The cross dimer or primer dimer check is an important design step to optimize primer performance in multiplex reactions. The primer specificity check is performed to minimize the risk of primer binding to human background DNA. The main improvement to existing oligonucleotide design web-tools is that oli2go combines multiple steps in an all-in-one solution, where other web applications only accomplish parts of the whole design workflow. See the GNU General Public License for more details. Candidates where the Tm is within the defined range are then checked for hairpin formation. It does not even involve as many sequence databases as oli2go for one run. Oli2go will output eligible primer pairs (each with one forward and reverse primer) that generate a product within the defined size range, do not form any secondary structures with each other, and show minimum difference in G values. In the most unlikely of these three scenarios, no primer dimers were identified after the first run of MFEprimer. As oligonucleotides are single stranded, they can form intramolecular secondary structures, such as hairpin loops, but also intermolecular structures, such as primer dimers (10). 16. Primer Suite can design primers for both native or bisulfite-treated DNA. The detection capability of the probe is dependent on the specificity of the associated primers and the preceding DNA amplification reaction. Multiplex bisulfite PCR resequencing is a scalable technique which can be used to analyse the methylation of multiple The cross dimer check using MFEprimer is another case where the results should be handled with care, as it is not as accurate as the cross dimer check provided with oli2go (Supplementary Table S2). Fragment Calculator Please enter your sequence information above and click the "CALCULATE" button. Acknowledgements A 45-plex assay targeting antibiotic resistance genes was successfully designed using oli2go and experimentally tested to evaluate the performance of the software. The oligo sequence is displayed in the Oligo Analysis Plot below where the full-length
1. again. As specificity checks are performed for each possible variable position, an increase in run time will be the consequence. The manual workflow was simulated by means of three different scenarios. None declared. Copyright (c) 2015 Bakhtiarizadeh etal. Although BLAST was not intended for specificity checks of oligonucleotides due to the lack of, e.g. The specific probes resulting from the preceding specificity check are used to find possible forward and reverse primer candidates that flank the hybridization oligonucleotide. A common list of parameters are used Primer Suite to design primers for both bisulfite or native primers. PrimerDimer Contact Us. However, the model needs to be selected carefully, as errors in Tm estimation may negatively influence the experiment outcome, e.g. The resulting alignment hits are compared to the target sequence hits generated in the file preparation workflow step. This means that primers with the same Tm can bind differently to the target due to different hybridization behaviors. Availability (2018), Sci. Tel: +43 66488256010; Email: NCBI database sources used for the probe specificity check, Oligonucleotide design by multilevel optimization, Bioinformatic tools and guideline for PCR primer design, Selection of primers for polymerase chain reaction, Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction, Versatility of different melting temperature (T, Primer3Plus, an enhanced web interface to Primer3, Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction, Physical principles and visual-OMP software for optimal PCR design, The thermodynamics of DNA structural motifs, PrecisePrimer: an easy-to-use web server for designing PCR primers for DNA library cloning and DNA shuffling, MFEprimer2.0: a fast thermodynamics-based program for checking PCR primer specificity, MSP-HTPrimer: a high-throughput primer design tool to improve assay design for DNA methylation analysis in epigenetics, A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics, An introduction to PCR primer design and optimization of amplification reactions, Optimization of the annealing temperature for DNA amplification in vitro, Fast and accurate short read alignment with BurrowsWheeler transform, Biopython: freely available Python tools for computational molecular biology and bioinformatics, Kraken: ultrafast metagenomic sequence classification using exact alignments, Sequencedependent thermodynamics of a coarse-grained DNA model, CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. References More precise algorithms using thermodynamic models or molecular dynamics simulations cannot handle the large number of possible oligonucleotide interactions in multiplex applications within a realistic time frame (16,23). The BLAST results comprise all hits that show >90% sequence similarity to the query sequence and form the basis for the specificity check of the probes. 7. Lu, J. Johnston, A.et al. This table can also be downloaded as comma-separated values (CSV) file. Oli2go represents an efficient, accurate, user friendly design tool for multiplex oligonucleotide projects. They are essential components in various applications such as polymerase chain reaction (PCR), microarrays, fluorescence in situ hybridization (FISH) and other DNA-based technologies. 13. It directly influences the prediction of Tm delivered by different calculation models. Conflict of interest statement. The data should include a minimum of two sequences, as oli2go is designed to handle more than one sequence for multiplex reactions. Rep. 10. Michaela Hendling, Stephan Pabinger, Konrad Peters, Noa Wolff, Rick Conzemius, Ivan Barii, Oli2go: an automated multiplex oligonucleotide design tool, Nucleic Acids Research, Volume 46, Issue W1, 2 July 2018, Pages W252W256, https://doi.org/10.1093/nar/gky319. o The PrimerROC interface is freely available at www.primer-dimer.com/roc. Even with these limitations in background data and thermodynamic alignments, the manual design workflow (Figure2) still showed worse performance than oli2go. PrimerROC Analysis for Dimer predictions PrimerPlex PrimerPlex Output If the sequence is suitable for your experiment, you can use the ADD OLIGO TO CART
Additionally, oli2go supports the option to generate two-part hybridization probes used in ligation-based experiments. Alternative approaches to improve the performance of the specificity check have been evaluated without success. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide, This PDF is available to Subscribers Only. Both cases should be avoided, as they may lead to false-negative signals in the experiment (10). PrimerDimer Output Dieffenbach C.W., Lowe T.M., Dveksler G.S. This step analyzes each possible probe candidate with BLAST against the user-defined databases (Table1). THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. Johnston, A. Lu, J. et al. If the results contain at least one primer pair for each sequence, each one is checked against the other primers in the results. Schematic illustration of the run time evaluation. 8. 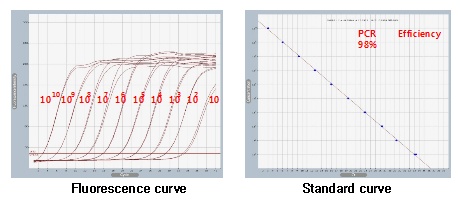 A 45-plex assay targeting the 45 most common antibiotic resistance genes was designed with oli2go as proof of principle. Only probes that bind to the same sequences as their target sequence, will be accepted. PrimerROC: accurate condition-independent dimer prediction using ROC analysis. To plot
A 45-plex assay targeting the 45 most common antibiotic resistance genes was designed with oli2go as proof of principle. Only probes that bind to the same sequences as their target sequence, will be accepted. PrimerROC: accurate condition-independent dimer prediction using ROC analysis. To plot
Sci. Properties of concatemer duplexes formed by d(T-G-C-A-C-A-T-G), Nucleotides.
The Fragment Calculator module is available at: http://www.primer-suite/fragcalc. The three scenarios differ by the number of required primer dimer checks. of the primer is compared against itself for self-hybridization potential. For this process, users are required to input an excel file with all the primers they wish to input with the following mandatory fields from the Primer Suite program: Results from PrimerPlex will be reported in an excel workbook. Download citation | Download article. applications is challenging due to unique limitations relating to bisulfite-converted template and multiplex reactions. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C, Remm M., Rozen S.G. Qu W., Zhou Y., Zhang Y., Lu Y., Wang X., Zhao D., Yang Y., Zhang C. Pandey R.V., Pulverer W., Kallmeyer R., Beikircher G., Pabinger S., Kriegner A., Weinhusel A. Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B.et al. 12. PrimerSuite String searching methods and data structures, such as suffix trees and Burrows-Wheeler-Transformation have been tested, but failed due to the high data amount of the background databases and the related memory usage (20). Inputs: This tool uses concentrations measured by 175 bp and 125 bp amplicons and the [175bp]/[125bp] ratio of these concentrations to estimate the average fragment length of a genomic or bisulfite-converted human DNA sample, the total number of genome copies in a measured sample, as well as the number of amplifiable (unbroken) instances of a DNA region of any length. Sequences containing ambiguous nucleotides are supported, but should be used carefully as each variable position within the sequence increases the number of computational steps. An overview of the oli2go software. PrimerSuite output Input sequences are first aligned using the standalone version of the National Center for Biotechnology Informations (NCBI) Basic Local Alignment Search Tool (BLAST) version 2.7.0+ and a comprehensive collection of databases (Table1). Designated input parameters are necessary for primer and probe design and dimerization checks. no or non-specific amplification of DNA duplexes (5). Your comment will be reviewed and published at the journal's discretion. Oli2go combines all essential steps for a high quality probe and primer design for a variety of biological experiments in an all-in-one solution. 3. your oligo against other sequences or against shorter versions of itself, please 7. 2. To date, a high number of web-tools for oligonucleotide designing are available (7,8,1215). The main output contains primers and probes for each input sequence in FASTA format. The exclamation mark indicates that certain input sequences result in more than 100 000 alignments making it almost impossible to examine all hits using online BLAST. In order to increase the possibility of achieving a specific amplification reaction, the sequence of the amplified product of the primers needs to be unique in comparison to other templates (2). PrimerDimer only looks for 'extension dimers' (dimers which has the potential to extend from the 3' end), which has been found to be the most problematic during our wet lab validations. These databases are a collection of sequence files covering >100 million sequences from bacteria, viruses, fungi, archaea, invertebrates, environmental samples, protozoa, plants and whole genome shotgun (WGS) projects, downloaded from the File Transfer Protocol (FTP) server of NCBI. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED.